Structure Comparison¶
This section shows how to find identical or similar protein chains in two PDB files and align them.
proteins module contains functions for matching and mapping
chains. Results can be used for RMSD fitting and PCA analysis.
Output will be AtomMap instances that can be used as input
to ProDy classes and functions.
Match chains¶
We start by importing everything from the ProDy package:
In [1]: from prody import *
In [2]: from pylab import *
In [3]: ion()
Matching chains is useful when comparing two structures. We will find matching chains in two different HIV Reverse Transcriptase structures.
First we define a function that prints information on paired (matched) chains:
In [4]: def printMatch(match):
...: print('Chain 1 : {}'.format(match[0]))
...: print('Chain 2 : {}'.format(match[1]))
...: print('Length : {}'.format(len(match[0])))
...: print('Seq identity: {}'.format(match[2]))
...: print('Seq overlap : {}'.format(match[3]))
...: print('RMSD : {}\n'.format(calcRMSD(match[0], match[1])))
...:
Now let’s parse bound RT structure 1vrt and unbound structure 1dlo:
In [5]: bound_all = parsePDB('1vrt')
In [6]: unbound_all = parsePDB('1dlo')
Let’s verify that these structures are not aligned:
In [7]: showProtein(unbound_all, bound_all);
In [8]: legend();
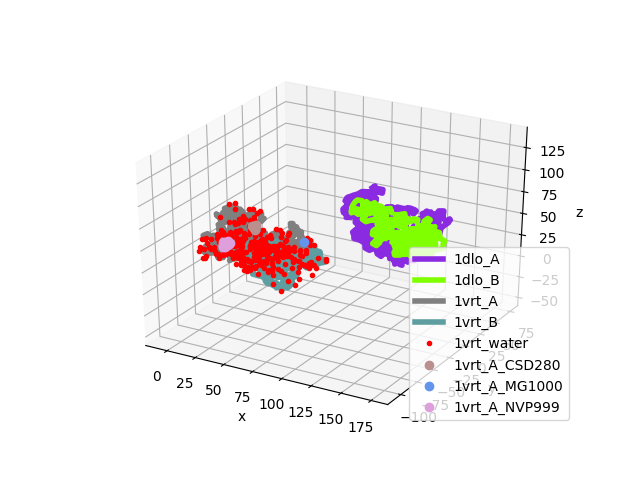
We find matching chains as follows:
We first select just the protein for matching:
In [9]: bound = bound_all.protein
In [10]: unbound = unbound_all.protein
In [11]: matches = matchChains(bound, unbound)
In [12]: for match in matches:
....: printMatch(match)
....:
Chain 1 : AtomMap Chain B from 1vrt -> Chain B from 1dlo
Chain 2 : AtomMap Chain B from 1dlo -> Chain B from 1vrt
Length : 400
Seq identity: 99.2518703242
Seq overlap : 96
RMSD : 110.45149192
Chain 1 : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Chain 2 : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Length : 524
Seq identity: 99.0458015267
Seq overlap : 94
RMSD : 142.084163869
This resulted in two matches. Chains A and B of two structures are paired. The chains in the matches contain only Cα atoms:
In [13]: match[0][0].iscalpha
Out[13]: True
In [14]: match[0][1].iscalpha
Out[14]: True
For a structural alignment based on both chains, we merge these matches as follows:
In [15]: bound_ca = matches[0][0] + matches[1][0]
In [16]: bound_ca
Out[16]: <AtomMap: (AtomMap Chain B from 1vrt -> Chain B from 1dlo) + (AtomMap Chain A from 1vrt -> Chain A from 1dlo) from 1vrt (924 atoms)>
In [17]: unbound_ca = matches[0][1] + matches[1][1]
In [18]: unbound_ca
Out[18]: <AtomMap: (AtomMap Chain B from 1dlo -> Chain B from 1vrt) + (AtomMap Chain A from 1dlo -> Chain A from 1vrt) from 1dlo (924 atoms)>
Let’s calculate RMSD:
In [19]: calcRMSD(bound_ca, unbound_ca)
Out[19]: 129.34348658001392
We find the transformation that minimizes RMSD between these two selections and apply it to unbound structure:
In [20]: calcTransformation(unbound_ca, bound_ca).apply(unbound);
In [21]: calcRMSD(bound_ca, unbound_ca)
Out[21]: 6.0020747465625375
Let’s see the aligned structures now:
In [22]: showProtein(unbound, bound);
In [23]: legend();
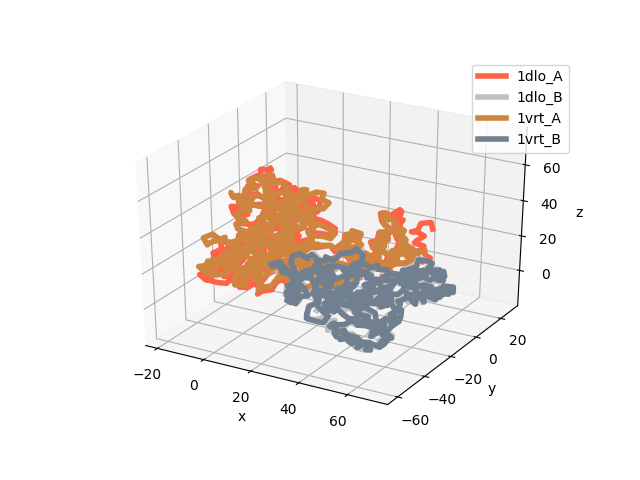
By default, matchChains() function matches Cα atoms.
subset argument allows for matching larger numbers of atoms.
We can match backbone atoms as follows:
In [24]: matches = matchChains(bound, unbound, subset='bb')
In [25]: for match in matches:
....: printMatch(match)
....:
Chain 1 : AtomMap Chain B from 1vrt -> Chain B from 1dlo
Chain 2 : AtomMap Chain B from 1dlo -> Chain B from 1vrt
Length : 1600
Seq identity: 99.2518703242
Seq overlap : 96
RMSD : 1.71102621571
Chain 1 : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Chain 2 : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Length : 2096
Seq identity: 99.0458015267
Seq overlap : 94
RMSD : 7.78386812028
Or, we can match all atoms as follows:
In [26]: matches = matchChains(bound, unbound, subset='all')
In [27]: for match in matches:
....: printMatch(match)
....:
Chain 1 : AtomMap Chain B from 1vrt -> Chain B from 1dlo
Chain 2 : AtomMap Chain B from 1dlo -> Chain B from 1vrt
Length : 3225
Seq identity: 99.2518703242
Seq overlap : 96
RMSD : 2.20947196284
Chain 1 : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Chain 2 : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Length : 4159
Seq identity: 99.0458015267
Seq overlap : 94
RMSD : 7.83814068858
Map onto a chain¶
Mapping is different from matching. When chains are matched, all matching
atoms are returned as AtomMap instances. When atoms
are mapped onto a chain, missing atoms are replaced by dummy atoms. The
length of the mapping is equal to the length of chain. Mapping is used
particularly useful in assembling coordinate data for the analysis of
heterogeneous datasets (see Ensemble Analysis).
Let’s map bound structure onto unbound chain A (subunit p66):
In [28]: def printMapping(mapping):
....: print('Mapped chain : {}'.format(mapping[0]))
....: print('Target chain : {}'.format(mapping[1]))
....: print('Mapping length : {}'.format(len(mapping[0])))
....: print('# of mapped atoms: {}'.format(mapping[0].numMapped()))
....: print('# of dummy atoms : {}'.format(mapping[0].numDummies()))
....: print('Sequence identity: {}'.format(mapping[2]))
....: print('Sequence overlap : {}\n'.format(mapping[3]))
....:
In [29]: unbound_hv = unbound.getHierView()
In [30]: unbound_A = unbound_hv['A']
In [31]: mappings = mapOntoChain(bound, unbound_A)
In [32]: for mapping in mappings:
....: printMapping(mapping)
....:
Mapped chain : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Mapping length : 4370
# of mapped atoms: 4159
# of dummy atoms : 211
Sequence identity: 99
Sequence overlap : 94
Mapped chain : AtomMap Chain B from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain B from 1vrt
Mapping length : 4370
# of mapped atoms: 3209
# of dummy atoms : 1161
Sequence identity: 99
Sequence overlap : 72
mapOntoChain() mapped all atoms. subset argument allows for
matching other sets of atoms. We can map backbone atoms as follows:
In [33]: mappings = mapOntoChain(bound, unbound_A, subset='bb')
In [34]: for mapping in mappings:
....: printMapping(mapping)
....:
Mapped chain : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Mapping length : 2224
# of mapped atoms: 2096
# of dummy atoms : 128
Sequence identity: 99
Sequence overlap : 94
Mapped chain : AtomMap Chain B from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain B from 1vrt
Mapping length : 2224
# of mapped atoms: 1604
# of dummy atoms : 620
Sequence identity: 99
Sequence overlap : 72
Or, we can map all atoms as follows:
In [35]: mappings = mapOntoChain(bound, unbound_A, subset='all')
In [36]: for mapping in mappings:
....: printMapping(mapping)
....:
Mapped chain : AtomMap Chain A from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain A from 1vrt
Mapping length : 4370
# of mapped atoms: 4159
# of dummy atoms : 211
Sequence identity: 99
Sequence overlap : 94
Mapped chain : AtomMap Chain B from 1vrt -> Chain A from 1dlo
Target chain : AtomMap Chain A from 1dlo -> Chain B from 1vrt
Mapping length : 4370
# of mapped atoms: 3209
# of dummy atoms : 1161
Sequence identity: 99
Sequence overlap : 72
